src.factorize_graph_matching.construct_sparse_aff_mat¶
- src.factorize_graph_matching.construct_sparse_aff_mat(Ke: torch.Tensor, Kp: torch.Tensor, row_idx: torch.Tensor, col_idx: torch.Tensor)[source]¶
Construct sparse affinity matrix with edge-wise affinity matrix \(\mathbf{K}_e\), node-wise matrix \(\mathbf{K}_p\) and graph connectivity matrices \(\mathbf{G}_1, \mathbf{H}_1, \mathbf{G}_2, \mathbf{H}_2\)
\[\mathbf{K}=\mathrm{diag}(\mathrm{vec}(\mathbf{K}_p)) + (\mathbf{G}_2 \otimes_{\mathcal{K}} \mathbf{G}_1) \mathrm{diag}(\mathrm{vec}(\mathbf{K}_e)) (\mathbf{H}_2 \otimes_{\mathcal{K}} \mathbf{H}_1)^\top\]where \(\mathrm{diag}(\cdot)\) means building a diagonal matrix based on the given vector, and \(\mathrm{vec}(\cdot)\) means column-wise vectorization. \(\otimes_{\mathcal{K}}\) denotes Kronecker product.
This function does not supports batched operations. This formulation is developed by “F. Zhou and F. Torre. Factorized Graph Matching. TPAMI 2015.”
- Parameters
Ke – \((1\times n_e)\) non-zero elements in edge-wise affinity matrix Ke. \(n_e\): number of non-zero elements in dense Ke
Kp – \((1\times n_1 n_2)\) non-zero elements in node-wise affinity matrix Kp. \(n_1\): number of nodes in graph 1, \(n_2\): number of nodes in graph 2
row_idx – \((1\times n_e)\) row indices of the non-zero elements in edge-wise affinity matrix Ke
col_idx – \((1\times n_e)\) column indices of the non-zero elements in edge-wise affinity matrix Ke
- Returns
none-zero values in the affinity matrix \(\mathbf K\), and their row/column indices (value, row-idx, col-idx)
For a graph matching problem with 5 nodes and 4 nodes, the connection of \(\mathbf K\) and \(\mathbf{K}_p, \mathbf{K}_e\) is illustrated as
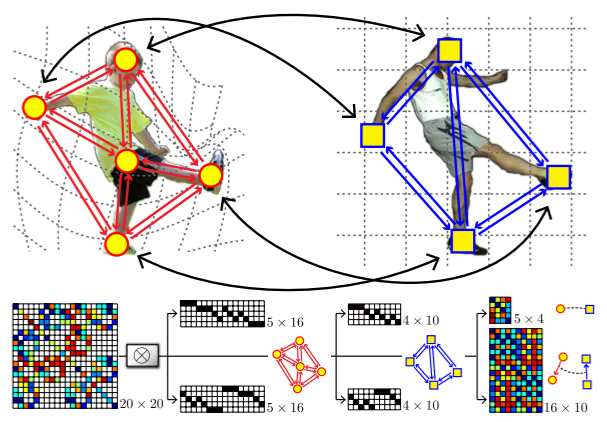
where \(\mathbf K (20 \times 20)\) is the complete affinity matrix, \(\mathbf{K}_p (5 \times 4)\) is the node-wise affinity matrix, \(\mathbf{K}_e(16 \times 10)\) is the edge-wise affinity matrix.